Science like sport has rules and procedures. One of the most important procedures is that if you want to claim something in science you have to devise an experiment and test the proposition. You then publish your procedures and your results so everyone can see what you did.
If people are happy with what you have done your claim is accepted. If they don’t like it they can repeat the work to check you haven’t made a mistake – or cheated. If they come up with the same results, your work and your claim is validated and accepted. If not then there’s a problem and you may be refuted.
These rules have the great merit of simplicity in theory and are supposed to ensure that no good work is lost because the procedure for verifying is so simple.
The story of Sasi and his side by side (SBS) alternative to the Watson Crick double helix (W-C) model of DNA is an example of these simple rules being broken and potentially critically important work being ignored.
Sasi and his supporters are free to make this accusation quite simply because his work was initially criticised and then dismissed/forgotten even though no one has ever bothered to repeat it in over 40 years.
The reasons for this are involved and speak to many different failings both institutional and individual in terms of peer pressure, herd mentality and serious problems with science publishing and funding. These things are simultaneously mundane and powerful. They are also thoroughly human.
If Sasi had been a junior researcher, if this work had been published in some obscure journals or had been examining some unimportant field it would perhaps matter less. But as you will see (Stokes Papers 14, p 224 para 3; and https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ (Stokes Papers 15b Ch II p 31-34) Sasi was at the time one of the very few people in the world qualified to undertake this work, the DNA molecule is one of the most important molecules known to science and his work was published in leading journals including Current Science, Nature, Proceedings of the National Academy of Science (PNAS) and Nucleic Acids Research.
It is effectively impossible to watch DNA in action, in vivo (in life). To view it we have to extract it from the cell. To do this we have to follow the procedures like those outlined on the DNA extraction procedure page on this https://sidebysidedna.com/johnson-materials/18-dna-extraction-procedure/ (Johnson Material 18). This is not subtle and by definition it strips away any and all associated material and leaves a cold and naked, in vitro (in glass/in the test tube) molecule, an artifact, created by human intervention. Then in addition the water content is reduced and the whole thing is crystallised, ‘frozen’. The thing you are left with is an in vitro artifact. It is a well understood principle in science that because they have been messed about with so much the relationship between any artifact and their in vivo versions has to be very carefully deduced/projected with lots of qualifications.
DNA is not and never has been a free entity in vivo but in order to understand it and work out its nature you have to strip it down and identify its range of movement in the same way as you might a human skeleton. This is a deliberately unsatisfactory analogy which we will flesh out later.
Even stripped down in vitro DNA is too small for us to observe directly. Our assessment of it is by those techniques including x ray analysis, diffraction patterns etc which are almost by necessity being done on a still/crystallised molecule.
If you are at a dance/disco and it is almost completely dark you know people are dancing but can’t say for sure exactly what moves they are making. Once the strobe light starts you get snapshots of those moves and from those you can infer the full arcs of their movement. This is also how movies work as a sequence of individual still frames.
This is effectively what Sasi and his team did. They generated a series of crystals of each element of the molecule and analysed those through different ranges of movement. It was from these that they generated the overall picture of the molecule.
This part of the team’s approach was not unique but what was unique was that they prepared much more complex crystals than had been used previously. They prepared samples of the bases from units that included the sugar phosphate backbone which made it far more realistic. Most importantly they did not limit their investigation by only examining right-handed options (supportive of the W-C) as most other groups previously had.
This aspect of their approach is what drew the attention of the philosophy of science student Terry Stokes and he did his PhD thesis on the role of logic in scientific innovation (part of the Popper/Kuhn debate). He used this group’s work as his worked example and in the process has left us a remarkable diary/analysis/character study of the key players in DNA research of the time. This thesis is reproduced in full https://sidebysidedna.com/stokes-papers/ Stokes Papers 15 PhD thesis) together with a shorter dissertation (Stokes Papers 14 Dissertation – unfortunately still unavailable on that site but possibly accessible to readers by request to publishers) both with Prof Stokes’ kind permission.
THE BACKBONE
Up until Sasi’s research all analysis of the secondary structure of the molecule had used mono nucleosides – very simple crystal preparations of the bases themselves. What Sasi had his team do was prepare and analyse di, tri and tetra nucleotide crystals, bases with backbone elements separating them. This meant that their analysis of the sugar phosphate ‘backbone’ included the effects of base stacking in creating the conformations. This allowed them to demonstrate the great range of viable options around the phosphodiester bonds in the sugar phosphate chains. (See https://sidebysidedna.com/sasisekharan-papers/is-3-nucleotide-rigid/ Sasisekharan Papers 8)
They showed that the deoxy ribonucleic ‘backbone’ of DNA has 4 low energy or stable positions with relatively low energy pathways between them on 3 sides. In effect the molecule can twist from position 1 to 2 to 3 to 4 and back again but it cannot go from 1 to 4 ie it can’t twist full circle, a bit like a human backbone.
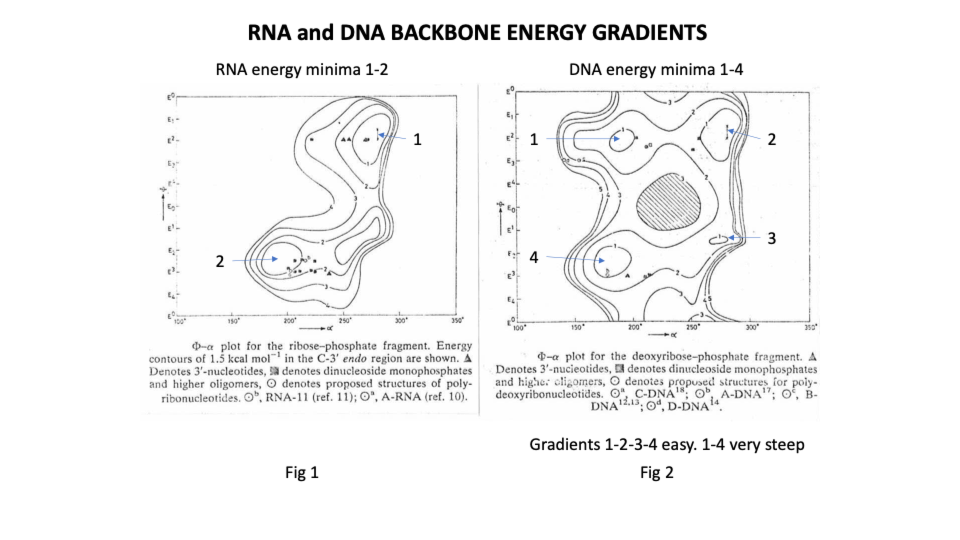
By contrast the ribonucleic backbone of RNA has only 2 stable positions because the extra hydroxyl group on C2 of the sugar restricts the rotation options of the phosphodiester bond (https://sidebysidedna.com/sasisekharan-papers/is-3-nucleotide-rigid/ Sasisekharan Papers 8 p 188).
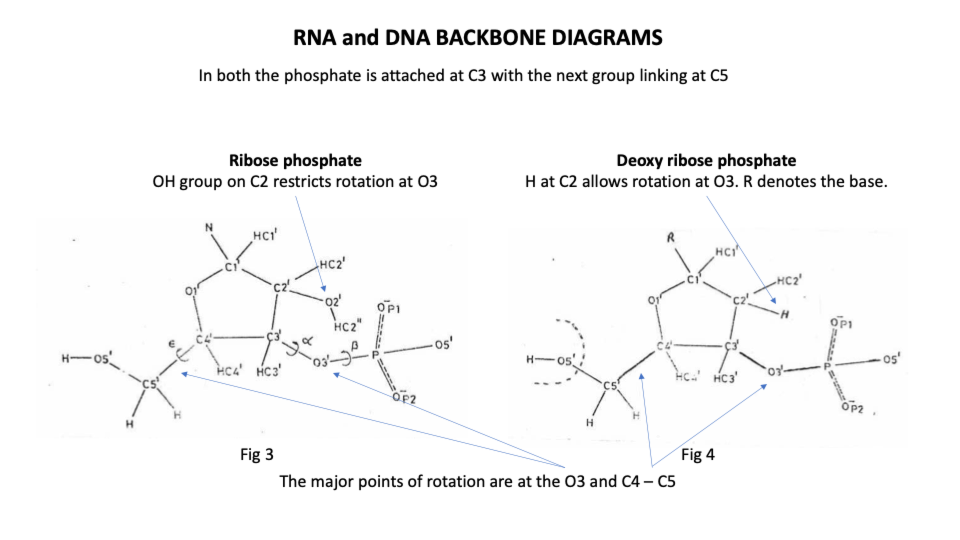
Non specialists may be surprised to learn that DNA is far more flexible than RNA which is simply not what the W-C suggests or even what we learn about tRNA ‘weaving’ its way through the cell.
We know that DNA emerged from the pre-existing ribonucleoprotein world (RNP) of which RNA, ribosomes and a few other molecules are retained functional remnants. The loss of its hydroxyl group made DNA capable of doing its job far more effectively than its RNP precursor.
Because of the absence of the extra hydroxyl group compared to RNA, DNA doesn’t make cross connections and so it maintains its overall shape no matter its sequence or length. DNA synthesis goes 50x quicker than RNA synthesis and protein synthesis.
UNWINDING: A MAJOR PROBLEM WITH THE WATSON CRICK DOUBLE HELIX
It is most unusual in nature for a molecule to be wrapped around itself like the W-C. This twisting makes it extremely difficult for it to disentangle itself. Disentangling either partially for template formation or fully for replication lies at the heart of DNA’s most critical functions.
The other thing that this makes very difficult is the binding of the histones. They need to find their binding sites by travelling round the molecule to the major groove. They then have to be out of the way during replication and relocate on daughter molecules.
The very important problem of unwinding was addressed head on by Crick and Watson in their 30 May 1953 follow up paper ‘Genetic Implications of the Structure of DNA’ Nature Vol 171 pp964-967 – see p966 para 4 where they write, ‘Although it is difficult at the moment to see how these processes occur without everything getting tangled, we do not feel that this objection will be insuperable.’
A very good history of this issue can be read in https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ (Stokes Papers 15b Ch III ‘The Unwinding Problem’ p 48).
Following similar work by other authors in 1975 the Polish biologist Gorski outlined the problem www.sidebysidedna.com Stokes Papers 14 p217 para 4 and https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ Stokes Papers 15b p51 para 4. The observed time it takes for the gut bacterium E.coli to replicate is between 30 and 41 minutes. With a single DNA strand of nearly 300,000 base pairs (2.82 x 105) which given the helical turn h of 10 for the molecule means the whole molecule would have to rotate at 6,900 – 9,000 revs/min or 115-150 revs/sec.
At the same time the associated histones etc would have to be delivered to the daughter strands which would themselves be connected to the still spinning mother molecule and would be rotating themselves as they took up their double helix form. We can picture a rapidly rotating ‘Y’ shaped molecule in an aqueous environment surrounded by delicate organelles, with highly polarised components attracting water molecules, creating drag etc.
In higher organisms there would be several DNA molecules performing this act of madness in close quarters simultaneously.
THE CHALLENGE IS TAKEN UP
This was the state of knowledge of DNA replication up until the mid 1970’s. Two decades after Watson and Crick themselves addressed this major problem there was no solution to it and yet no one had done a coherent study of the molecule and still less seriously examined an alternative model.
This is where Sasi and another, more theoretical group in New Zealand came in proposing the SBS. (www.sidebysidedna.com Stokes Papers 14 p 222 para 2 and p 224 para 3 and https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ Stokes Papers 15b Ch I NZ and Ch II Indian). The point of the SBS model being that the unwinding is simply not an issue. There is no unwinding to be done because the two chains lie side by side and are simply pulled apart.
In addition the histone bonding is very straightforward on such an open structure as is histone release during template formation and replication.
In 1970-71 Sasi was Visiting Professor at Princeton working alongside Rob Langridge who had done the first study of base stacking in 1960. This and subsequent papers by Pullman in 1962 and De Voe in 1968 all assessed right-handed stacking only, the others following Langridge’s lead.
In 1970 the Matsui research group observed from both X-ray and circular dichroism (CD) data that the synthetic D-DNA was left-handed. They couldn’t convert it into A-DNA or B-DNA, regarded as the most similar to naturally occurring forms.
By doing more careful and detailed work than the Matsui group Sasi showed that D-DNA could be converted to A-DNA and B-DNA which were evidently right-handed by both X-ray and CD data. Because he was able to do this by simply changing the humidity and such a simple change could not be responsible for something as profound as a change in helical sense he concluded that all 3 forms were right handed.
However the anomaly of the CD data indicating that D-DNA was left-handed remained. Sasi took up this issue with Langridge. He also raised the broader point about why in his original 1960 paper Langridge hadn’t also investigated left handed base stacking options. Sasi describes Langridge, who had been one of the Matsui researchers as being ‘evasive’ on these points. (other site Stokes Papers 14 p225 and https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ Stokes Papers 15b ChII p32 para 39).
This may be because of the implied criticism of the Matsui group’s methods or else as Sasi acknowledges because Langridge was distracted by plans to set up his new lab in San Francisco.
The observation of the anomaly and of the wider imprecision of much of the other work on DNA structure lead Sasi to a take a very significant decision. He left Princeton to take up his position as head of the Molecular Biophysics Unit at the Indian Institute of Science in Bangalore. As head of department he would be able to initiate his own research and he decided to examine all the component elements of the DNA molecule in fine and accurate detail.
The big problem was that he could recruit no molecular biologists or biochemists as doctoral research assistants. This was because he was proposing to examine left-handed possibilities and they viewed this as too unorthodox and career threatening to get involved with. (www.sidebysidedna.com Stokes Papers 14 p 226 para 2 and https://sidebysidedna.com/stokes-papers/terry-stokes-phd-chapters-i-v/ Stokes Papers 15b Ch II p 35)
Finally after 2 years he recruited N. Pattabiraman and G. Gupta, a nuclear physicist and particle physicist respectively, one of whom had never heard of DNA. This ignorance was an advantage. They had no preconceptions and were very good mathematicians.
These three were the core research team responsible for the initial set of key papers published in Current Science, Nature, PNAS and Nucleic Acids Research from 1976 to 1980. ( https://sidebysidedna.com/sasisekharan-papers/ Sasisekharan Papers 1-8).
THE CORE FINDINGS
Sasi’s team found that there was great flexibility in the sugar phosphate backbone. Like a regular chain it had strong covalently bonded links running along its length but that like a regular chain it had great flexibility between links ( https://sidebysidedna.com/sasisekharan-papers/ Sasisekharan Papers 1 and 2). They found that the bases bond covalently to the links and then bond to each other by relatively weak hydrogen bonding, holding the two chains together. This much is pretty much agreed between SBS and W-C. Sasi also agreed that the lowest energy stacking angle was 35o.
The departure for Sasi was that the lowest energy base stacking could be 35o for right or left-handed in most cases and that there are viable energy gradients between the two stacks again in most cases. Their two papers in Nucleic Acids Research ( https://sidebysidedna.com/sasisekharan-papers/ Sasisekharan Papers 3 and 4) shows these sequence specific stacking energy profiles.
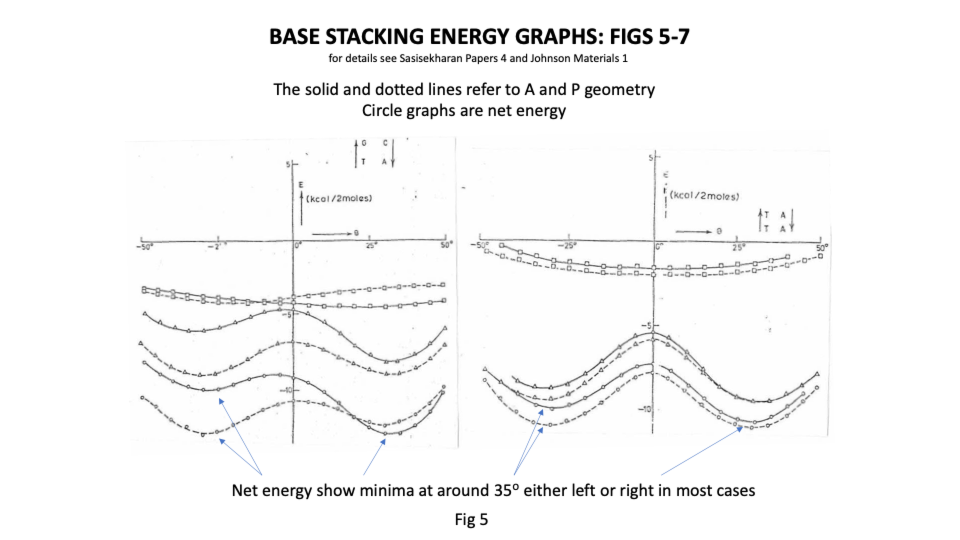
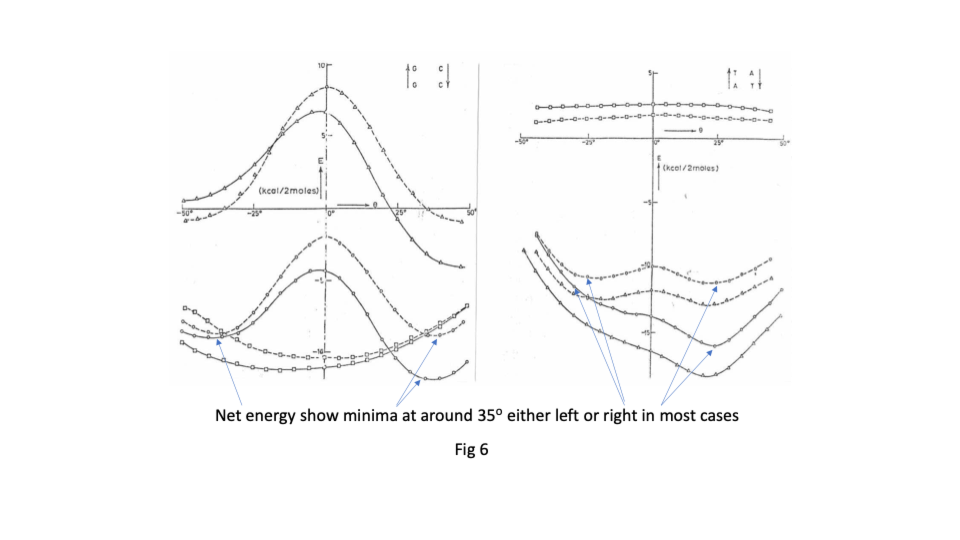
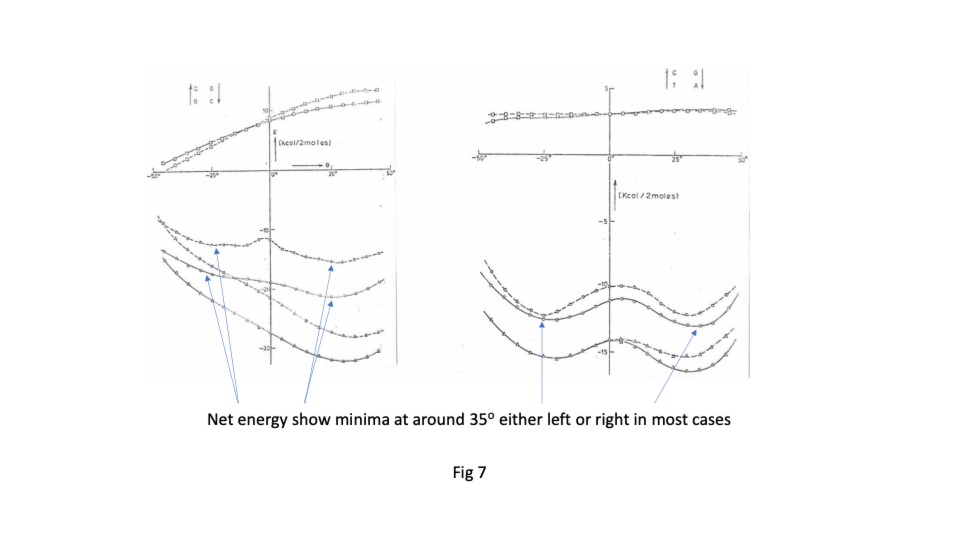
What this implies is that the tertiary structure of the molecule is determined by the stacking between the bases and so the tertiary structure of the molecule is sequence specific. ( https://sidebysidedna.com/sasisekharan-papers/ Sasisekharan Papers 5-10).
INDEPENDENT CORROBORATION? Another left-handed form appears
The front cover of Nature Vol 282 Issue 5740 published on 13th December 1979 ( https://sidebysidedna.com/johnson-materials/16-dna-is-not-always-a-simple-double-helix/ Johnson Material 16, p 28 and online search) heralded the discovery of the ‘only’ left-handed DNA known to science. This was the famous Zig Zag which had been discovered by a group at MIT headed by Alex Rich. This model is still a double helix but clearly shows that left-handed elements are viable and significant.
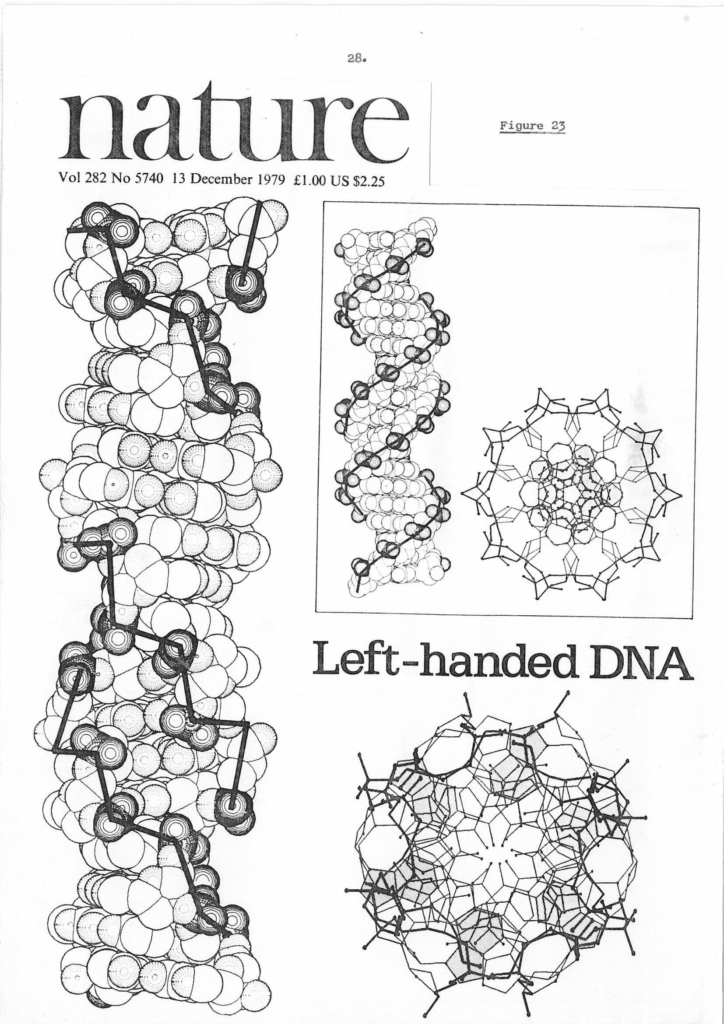
Given that Sasi had published a paper outlining the parameters of left and right-handed stacking in Nature the year before ( https://sidebysidedna.com/sasisekharan-papers/structure-of-dna-predicted-from-stereochemistry-of-nucleoside-derivatives/ Sasisekharan Papers 6) with references to all his previous papers it is very strange from a science procedure/rules and publishing point of view that Rich and his team should claim that their model was the ‘only’ left handed example known to science. They do reference Sasi’s 1978 PNAS paper ( https://sidebysidedna.com/sasisekharan-papers/some-implications-of-an-alternative-structure-for-dna/ Sasisekharan Papers 5) but still claim uniqueness. It is also strange that the referees and then the editor of Nature, David Davies indulged their claim.
Sasi’s work is a comprehensive analysis of all the component elements of the molecule whereas the MIT work is a one off. You can see Sasi’s remarkable account of the background to the MIT announcement in the letter written to A Johnson in February 1982 (https://sidebysidedna.com/sasisekharan-papers/letter-to-a-johnson/ Sasisekharan Papers 11) and this site https://singingdancingdna.com/letter-to-a-johnson/ .
What is so interesting is that the computer the MIT team were using had no preconceptions about DNA structure either and despite their best efforts to ‘make’ it support the W-C the computer forced them to realise their mistake. In this instance the computer saved the scientists from their own lack of scientific objectivity – which is very much a theme of this story.
(Up until the Zig Zag paper one authority had even taken to saying that left-handed DNA couldn’t exist because the ribose sugars are in the dextro rather than laevo chiral form. This is like saying cars with the steering wheel on the left can’t make right hand turns. There is this sense of people trying much too hard.)
What is very surprising is that this independent support for such a significant part of Sasi’s work didn’t lead to his work being re-evaluated or repeated. Part of this has to do with a shift in science publishing which impacted science practice in profound and unhelpful ways from the mid 1970’s.
This shift is well described in this article by science journalist Stephen Buranyi in the UK’s Guardian newspaper on 27 June 2017 ‘Is the staggeringly profitable business of scientific publishing bad for science?’. https://www.theguardian.com/science/2017/jun/27/profitable-business-scientific-publishing-bad-for-science. He outlines the post WWII history of science publishing which has led to a system described by many scientists and observers as ‘corrupt’.
That can be argued but what is certainly true is that the changes in publishing dis incentivised researchers from repeating/proving other people’s work. This disrupted the important verification/refutation process which is a core part of science rules and procedures. This is obviously an institutional problem but is very recognisably human. It is very damaging for science in general and in this particular case it is critical.
The ascription of the Zig Zag DNA as a ‘high salt’ anomaly and the ‘only’ left-handed version of the molecule became the received wisdom. It is deeply problematic and the failure to carefully examine this left-handed ‘anomaly’ and consider the wider implications of it is a glaring failure of science procedure/rules.
The way this fudge was so easily accepted makes about as much sense as when people say, ‘All xxxxx’s are a bit dumb but I have a friend who is an xxxxx but she/he is different.’ This is a standard way for human beings to have dodgy attitudes but maintain their human decency by celebrating their ability to make genuine friendships despite them. It’s a cake and eat it thing and has no place in the world as a whole and still less in science.
This loopy ‘high salt anomaly’ explanation of the ‘Zig Zag’ was for over a decade included as a footnote in the DNA section of the print edition of Encyclopaedia Britannica for instance. The author of that section on DNA was….Alex Rich.
Interestingly the only person to actually look systematically into the effects of changing salt concentrations on the left/right structure of DNA was Sasi. Working with new team members he produced papers in 1981 and 1984 which show that these changes are reversible ( https://sidebysidedna.com/sasisekharan-papers/b-to-z-transition-in-dna-fibre-the-question-of-handedness-of-the-duplex/ Sasisekharan Papers 9 and 10).
Again the fact that these supposedly profound structural changes are so easily reversible should have rung important alarm bells and garnered serious attention. But to science’s shame and detriment it didn’t.